A two-sample Mendelian randomized study of causality between lncRNAs and lung adenocarcinoma
-
摘要:
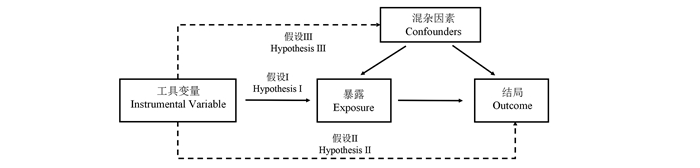
目的 利用两样本孟德尔随机化方法探究长链非编码RNAs(long-chain non-coding RNAs, lncRNAs)与肺腺癌发病风险之间的因果关联。 方法 采用肺腺癌全基因组关联分析数据和eQTL Gene联盟的cis-eQTL数据集,将与肺腺癌密切相关的单核苷酸多态性(single nucleotide polymorphisms, SNPs)作为工具变量,运用逆方差加权法、MR-Egger回归模型、加权中位数分析法、简单模式法和加权模式法5种两样本孟德尔随机化(Mendelian randomization, MR)模型来评估lncRNAs与肺腺癌之间的因果效应,并进行异质性检验、基因多效性检验和敏感性分析来评估结果的可靠性和稳定性。 结果 PVT1、LINC00824、Z94721.1与肺腺癌的关联效应值差异有统计学意义(均P<0.05),PVT1(与肺腺癌的效应值OR=0.79,95% CI:0.71~0.89)、LINC00824(与肺腺癌的效应值OR=0.59,95% CI:0.42~0.83)降低了原发性支气管肺癌(肺癌)的发病风险;而Z94721.1(与肺腺癌的效应值OR=1.09,95% CI:1.03~1.16)增加肺癌的发病风险,并且均通过了异质性检验、基因多效性检验和敏感性分析。 结论 PVT1、LINC00824、Z94721.1 3个lncRNAs与肺腺癌之间存在稳定的因果关联,为肺腺癌发病机制的研究提供了重要理论依据。 Abstract:Objective A two-sample Mendelian randomization method was used to investigate the causal relationship between long-chain non-coding RNAs(lncRNAs) and the risk of lung adenocarcinoma. Methods The genome-wide association study data of lung adenocarcinoma and the cis-eQTL data set of eQTL Gene Alliance were used. The single nucleotide polymorphisms (SNPs) closely related to lung adenocarcinoma was used as a tool variable. Five two-sample Mendelian randomization models, including the inverse variance weighting method, MR-Egger regression model, weighted median analysis method, simple mode method and weighted mode method, were applied to evaluate the causal effect between lncRNAs and the risk of lung adenocarcinoma. Moreover, the heterogeneity test, gene pleiotropy test and sensitivity analysis were carried out to evaluate the reliability and stability of the results. Results PVT1, LINC00824, Z94721.1 exhibited significant association with lung adenocarcinoma (all P < 0.05). The OR was 0.79 (95% CI: 0.71-0.89) for PVT1 and 0.59 (95% CI: 0.42-0.83) for LINC00824, indicating that both PVT1 and LINC00824 reduced the risk of lung cancer. The OR was 1.09 (95% CI: 1.03-1.16) for Z94721.1, showing that Z94721.1 increased the risk of lung cancer. All passed the heterogeneity test, gene pleiotropy test and sensitivity analysis. Conclusions The three lncRNAs, PVT1, LINC00824 and Z94721.1, have stable causal association with lung adenocarcinoma, providing an important theoretical basis for the study of the pathogenesis of lung adenocarcinoma. -
图 2 3个lncRNA的5种MR模型散点图
MR:孟德尔随机化;IVW:逆方差加权法;MR Egger:MR-Egger回归;SM:简单模式法;WME: 加权中位数法;WM:加权模式法;LUAD:肺腺癌;SNPs:单核苷酸多态性。
Figure 2. Scatter plots of 5 mendelian randomization models
MR: Mendelian randomization; IVW: inverse variance weighted; MR Egger: mendelian randomization Egger regression; SM: simple mode; WME: weighted median estimator; WM: weighted mode; LUAD: lung adenocarcinoma SNPs: single nucleotide polymorphisms.
表 1 lncRNAs的MR分析效应值结果
Table 1. Mendelian randomization estimates of lncRNAs
lncRNAs Ensembl ID MR法MR method SNPs β sx OR值value (95% CI) P值value PVT1 ENSG00000249859 IVW 26 -0.23 0.06 0.79(0.71~0.89) 4.22×10-5 LINC00824 ENSG00000254275 IVW 3 -0.53 0.18 0.59(0.42~0.83) 2.89×10-3 Z94721.1 ENSG00000227598 IVW 14 0.09 0.03 1.09(1.03~1.16) 4.08×10-3 LINC01891 ENSG00000231682 IVW 22 0.03 0.03 1.03(0.98~1.10) 2.56×10-1 LINC00511 ENSG00000227036 IVW 14 -0.02 0.04 0.98(0.91~1.06) 5.82×10-1 AC092142.1 ENSG00000261235 Wald Ratio 1 -0.34 0.44 0.71(0.30~1.67) 4.37×10-1 注:lncRNAs:长链非编码RNAs; MR: 孟德尔随机化; IVW,逆方差加权法;SNPs,单核苷酸多态性。
Notes: lncRNAs:long non-coding RNAs; MR: Mendelian randomization; IVW, inverse variance weighted; SNPs, single nucleotide polymorphisms.表 2 lncRNAs的MR分析效应值结果
Table 2. Mendelian randomization estimates of lncRNAs
lncRNAs Ensemble ID MR法MR method β sx OR值value (95% CI) P值value P异质性检验
PheterogeneityP多效性检验
PpleiotropyPVT1 ENSG00000249859 MR Egger -0.34 0.10 0.71(0.58~0.87) 3.24×10-3 0.467 0.245 WME -0.24 0.08 0.79(0.68~0.92) 2.26×10-3 IVW -0.23 0.06 0.79(0.71~0.89) 4.22×10-5 0.444 - 简单模式法Simple mode -0.25 0.14 0.78(0.60~1.02) 7.84×10-2 加权模式法Weighted mode -0.23 0.08 0.79(0.68~0.92) 6.58×10-3 LINC00824 ENSG00000254275 MR Egger -0.35 0.65 0.70(0.20~2.51) 6.85×10-1 0.392 0.821 WME -0.47 0.20 0.62(0.42~0.93) 2.07×10-2 IVW -0.53 0.18 0.59(0.42~0.83) 2.89×10-3 0.665 - 简单模式法Simple mode -0.42 0.25 0.65(0.40~1.07) 2.32×10-1 加权模式法Weighted mode -0.44 0.25 0.64(0.40~1.04) 2.17×10-1 Z94721.1 ENSG00000227598 MR Egger 0.02 0.06 1.02(0.91~1.14) 7.53×10-1 0.868 0.175 WME 0.08 0.04 1.08(1.00~1.18) 5.05×10-2 IVW 0.09 0.03 1.09(1.03~1.16) 4.08×10-3 0.779 - 简单模式法Simple mode 0.07 0.07 1.07(0.93~1.23) 3.70×10-1 加权模式法Weighted mode 0.07 0.05 1.07(0.97~1.17) 1.82×10-1 注:1. lncRNAs:长链非编码RNAs; MR: 孟德尔随机化; MR Egger,MR-Egger回归;WME,加权中位数法;IVW,逆方差加权法。
2. “-”表示无数值。
Notes: 1. lncRNAs:long non-coding RNAs; MR: Mendelian randomization; MR Egger, Mendelian randomization Egger regression; WME, weighted median estimator; IVW, inverse variance weighted.
2. "-" represents an infinite number of value. -
[1] Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71(3): 209-249. DOI: 10.3322/caac.21660. [2] Hutchinson BD, Shroff GS, Truong MT, et al. Spectrum of lung adenocarcinoma[J]. Semin Ultrasound CT MR, 2019, 40(3): 255-264. DOI: 10.1053/j.sult.2018.11.009. [3] 王宇松, 吉晓莹, 穆楠, 等. 非小细胞肺癌分子靶向治疗标志物研究进展[J]. 中国癌症防治杂志, 2020, 12(3): 285-290. DOI: 10.3969/j.issn.1674-5671.2020.03.09.Wang YS, Ji XY, Mu N, et al. Research progress of molecular targeted therapy markers for non-small cell lung cancer[J]. Chin J Oncol Prev Treat, 2020, 12(3): 285-290. DOI: 10.3969/j.issn.1674-5671.2020.03.09. [4] Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes[J]. Science, 2010, 329(5992): 689-693. DOI: 10.1126/science.1192002. [5] Feng C, Zhao Y, Li Y, et al. LncRNA MALAT1 promotes lung cancer proliferation and gefitinib resistance by acting as a miR-200a sponge[J]. Arch Bronconeumol (Engl Ed), 2019, 55(12): 627-633. DOI: 10.1016/j.arbres.2019.03.026. [6] Smith GD, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations[J]. Int J Epidemiol, 2004, 33(1): 30-42. DOI: 10.1093/ije/dyh132. [7] 王莉娜, Zhang Zuofeng. 孟德尔随机化法在因果推断中的应用[J]. 中华流行病学杂志, 2017, 38(4): 547-552. DOI: 10.3760/cma.j.issn.0254-6450.2017.04.027.Wang LN, Zhang ZF. Mendelian randomization approach, used for causal inferences[J]. Chin J Epidemiol, 2017, 38(4): 547-552. DOI: 10.3760/cma.j.issn.0254-6450.2017.04.027. [8] Wang Y, McKay JD, Rafnar T, et al. Corrigendum: Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer[J]. Nat Genet, 2017, 49(4): 651. DOI: 10.1038/ng0417-651a. [9] Horsfall LJ, Burgess S, Hall I, et al. Genetically raised serum bilirubin levels and lung cancer: a cohort study and Mendelian randomisation using UK Biobank[J]. Thorax, 2020, 75(11): 955-964. DOI: 10.1136/thoraxjnl-2020-214756. [10] Võsa U, Claringbould A, Westra HJ, et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression[J]. Nat Genet, 2021, 53(9): 1300-1310. DOI: 10.1038/s41588-021-00913-z. [11] Sanderson E, Davey Smith G, Windmeijer F, et al. An examination of multivariable Mendelian randomization in the single-sample and two-sample summary data settings[J]. Int J Epidemiol, 2019, 48(3): 713-727. DOI: 10.1093/ije/dyy262. [12] Bowden J, Holmes MV. Meta-analysis and Mendelian randomization: a review[J]. Res Synth Methods. 2019, 10(4): 486-496. DOI: 10.1002/jrsm.1346. [13] Hartwig FP, Davey SG, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption[J]. Int J Epidemiol, 2017, 46(6): 1985-1998. DOI: 10.1093/ije/dyx102. [14] Lawlor DA, Harbord RM, Sterne JA, et al. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology[J]. Stat Medi, 2008, 27(8): 1133-1163. DOI: 10.1002/sim.3034. [15] Zhou W, Liu G, Hung RJ, et al. Causal relationships between body mass index, smoking and lung cancer: Univariable and multivariable Mendelian randomization[J]. Int J Cancer, 2021, 148(5): 1077-1086. DOI: 10.1002/ijc.33292. [16] Wong J, Zhang H, Hsiung CA, et al. Tuberculosis infection and lung adenocarcinoma: Mendelian randomization and pathway analysis of genome-wide association study data from never-smoking Asian women[J]. Genomics, 2020, 112(2): 1223-1232. DOI:10. 1016/j.ygeno. 2019.07.008. [17] Zhang Z, Li H, Li J, et al. Polymorphisms in the PVT1 gene and susceptibility to the lung cancer in a Chinese northeast population: a case-control study[J]. J Cancer, 2020, 11(2): 468-478. DOI: 10.7150/jca.34320. [18] Zhou X, Zhang Y, Zhang Y, et al. LINC01414/LINC00824 genetic polymorphisms in association with the susceptibility of chronic obstructive pulmonary disease[J]. BMC Pulm Med, 2021, 21(1): 213. DOI: 10.1186/s12890-021-01579-3. -





 下载:
下载: