rs12252 polymorphism of IFITM3 gene and influenza susceptibility in Chinese population: a study-based sequential Meta-analysis
-
摘要:
目的 系统评价干扰素诱导跨膜蛋白3(interferon-induced transmembrane protein 3,IFITM3)基因rs12252位点多态性与中国人群流感易感性的定量关系。 方法 检索Medline、PubMed、Embase、Web of Science以及中国知网、万方、维普数据库,收集IFITM3与中国人群流感易感性相关的文章。采用Meta分析方法对收集的文章进行定量地综合分析。 结果 本次研究共纳入7篇文献,共计919例流感感染的病例,涉及到甲型流感(H7N9、H1N1pmd09、H3N2)及乙型流感病毒,序贯Meta分析显示本研究纳入的研究总样本已达到了得到稳定阳性结局所需的样本量。Meta分析结果发现,IFITM3基因的rs12252多态性与中国人群流感病毒易感性存在关联,携带rs12252 C等位基因的人群更易发生流感(C vs T:OR=1.67,95% CI:1.45~1.92;CC vs TT:OR=2.61,95% CI:1.97~3.46;TC vs TT:OR=1.55,95% CI:1.20~2.00;CC vs TC+TT:OR=2.01,95% CI:1.49~2.72;CC+TC vs TT:OR=8.90,95% CI:4.94~16.06)。 结论 IFITM3基因rs12252位点多态性是中国人群发生流感的危险因素。 -
关键词:
- IFITM3 /
- rs12252位点多态性 /
- 流感 /
- Meta分析 /
Abstract:Objective To systematically and quantitatively evaluate the relationship of rs12252 polymorphism in IFITM3 gene and susceptibility to influenza in Chinese population. Methods The databases of Medline, PubMed, Embase, Web of Science, China National knowledge infrastructure (CNKI), Wanfang and VIP were searched to retrieve the articles which focused on the topic of relationship between the rs12252 polymorphism and susceptibility to influenza in Chinese population. Meta-analysis method was used to comprehensively and quantitatively analyze the enrolled articles. Results Seven papers with a total of 919 influenza cases were involved in this article. Influenza virus involved influenza A (H7N9, H1N1 pmd09, H3N2) and influenza B. Study-based sequential meta-analysis of the study showed that the total sample size of this study have achieved the required sample size to obtain stable positive results. The results of this meta-analysis showed that the rs12252 polymorphism was associated with susceptibility to influenza in Chinese population and that the rs12252C polymorphism in IFITM3 gene was more susceptible to have influenza in Chinese people (C vs T: OR=1.67, 95% CI: 1.45-1.92; CC vs TT: OR=2.61, 95% CI: 1.97-3.46; TC vs TT: OR=1.55, 95% CI: 1.20-2.00; CC vs TC+TT: OR=2.01, 95% CI: 1.49-2.72; CC+TC vs TT: OR=8.90, 95% CI: 4.94-16.06). Conclusion the rs12252 polymorphism of IFITM3 gene was a risk factor for influenza in Chinese population. -
Key words:
- IFITM3 /
- rs12252 /
- Polymorphism /
- Influenza /
- Meta-analysis
-
流行性感冒(流感)是由流感病毒引起的一种急性呼吸道传染病,可导致住院甚至是死亡,对公众健康构成严重的威胁。据世界卫生组织统计显示,每年的流感流行将导致300~500万重症病例,大约25~50万死亡病例发生[1]。
流感的发生发展是病毒和宿主因素相互作用的结果,已有研究[2-3]证实了宿主因素对流感病毒引发疾病有着关键的作用。目前,越来越多的动物实验和基于人群的临床实验均证实,宿主基因的变异与宿主感染流感的易感性有关。干扰素诱导跨膜蛋白3(interferon-induced transmembrane protein 3,IFITM3)基因一直以来是研究的热点。IFITM3是一类宿主限制因子,具有抗病毒的活性。IFITM3的抗病毒效应是通过破坏细胞内胆固醇稳态来阻止流感病毒进入宿主细胞[4]。一方面它可以阻断病毒-内小体半融合后融合孔的形成[5],另一方面小鼠实验表明IFITM3还能促进肺内CD8 T细胞的存活,有助于清除病毒感染[6]。研究[7]发现在2009年H1N1流感季节性或大流行住院患者中,IFITM3等位基因(SNP rs12252 -C)富集,改变了流感病毒剪接受体位点。其原因主要是IFITM3多态rs12252 -C编码一种异常形式的IFITM3,即氨基酸端(N端)缺乏21个氨基酸(Δ21 IFITM3),Δ21 IFITM3通过阻断病毒复制的早期阶段来降低细胞对流感病毒的抵抗力[8-9]。
IFITM3基因rs12252位点多态性与流感病毒感染的关系成为近年来的研究热点。2016年以来有多项研究报道该方向的结果,结论仍然存在争议。多数研究的结论认为IFITM3基因rs12252位点基因多态性与流感病毒感染存在关联,这些研究[10-12]的人群主要集中在亚洲人群,尤其是中国人群。而在欧洲人群中几乎都是未找到存在关联的证据[8, 13-16],原因可能是欧洲人群rs12252是低频突变[17]。因此,本研究拟采用Meta分析的方法,综合定量分析不同的研究结果,系统的评价中国人群流感易感性与IFITM3基因rs12252位点多态性的关系。
1. 方法与材料
1.1 文献检索与纳入
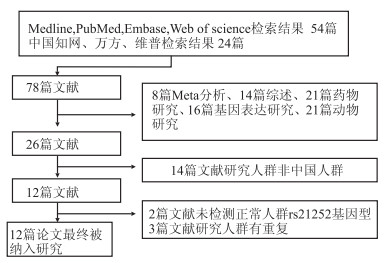
本研究的主题为IFITM3基因rs12252与中国人群流感病毒感染易感性的关系。本文检索了Medline、PubMed、Embase、Web of Science以及中国知网、万方、维普数据库(最后检索更新时间是2018年10月1日),检索策略为“流感”或“季节性流感”或“禽流感”或“H7N9”或“甲型流感”或“乙型流感”、“宿主基因”、“易感性”、“多态性”、“干扰素诱导跨膜蛋白基因”或“ rs12252 ”或“IFITM”。本次研究仅纳入报告数据可供二次分析的原始研究,对论文的语言和发表形式不做限制。当同一病例研究群体中被多次研究且多次发表时,将根据实际情况经两名研究者讨论后纳入其中质量最高的研究。图 1为本研究文献纳入排除的流程图。
1.2 文献纳入与排除标准及质量评价
1.2.1 文献纳入标准
(1) Meta分析所需的研究数据可以从原文全文或摘要中获取,或可以通过联系研究作者获取;(2)研究方式为病例对照研究或者队列研究;(3)所研究人群均为中国人群。
1.2.2 文献排除标准
(1) 无流感人群组或无正常对照人群;(2)重复发表;(3)研究中的数据不可用;(3)联系原文作者未回应或不能提供可供Meta分析的数据;(4)研究对象为非中国人群。
1.2.3 质量评价
由于纳入的研究均为病例对照研究,因此我们采用Newcastle-Ottawa Scale (NOS)非随机研究的质量评价量表中的病例对照研究分量表评价文献质量。该量表主要从研究对象的选择、暴露的评估及组间可比性三个方面对研究的质量进行评价。(NOS非随机研究的质量评价量表英文版介绍网址: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp)
1.3 统计学方法
由课题组双人提取和录入文献数据,如遇问题再请第三人共同讨论后决定。研究的数据处理采用R语言分析,统计分析包为Meta v 4.9-2 (作者Guido Schwarzer)。由于本次研究纳入的文献均为病例对照研究设计,故对总体的合并效应值采用比值比(OR)及95%的置信区间(95% CI),检验水准α=0.05。异质性的大小采用I2值来估计,并采用Q检验,检验水准α=0.1。当存在异质性时,采用随机效应模型,否则采用固定效应模型。发表偏倚采用Egger线性回归法检验,检验水准α=0.1。敏感性分析采用剪补法来估计,分别观察剔除某一研究后效应值的变化是否显著改变。本研究方法的样本含量估计采用试验序贯分析的理论,计算方法参考Jørn Wetterslev的报道,软件分析采用TSA v0.9软件(哥本哈根临床试验中心(Copenhagen trial unit,CTU))[18]。
2. 结果
2.1 纳入研究基本信息及质量评价
本次研究共纳入7篇符合上述排除标准的文献,共计919例流感感染的病例,涉及到甲型流感(H7N9、H1N1pmd09、H3N2)及乙型流感病毒,其中2篇是在儿童中开展的研究,3篇在全年龄组中开展的研究,2篇在成人中开展的研究,见表 1。
表 1 研究纳入文献的基本信息Table 1. Basic information of the included studies作者 发表年 研究对象 流感型别 流感病例基因分型 正常人群基因分型 TT TC CC TT TC CC Jianbing Zheng[19] 2016 儿童 未分型 6 25 29 49 98 50 Lee[10] 2017 成人 H7N9 11 25 15 28 48 29 H1N1pmd09 51 90 83 28 48 29 Qiaowei Zhu[20] 2013 儿童 Influenza 6 29 9 73 72 52 Yang Pan[21] 2016 全年龄组 H3N2 8 45 53 16 34 15 Yamagata 10 19 24 16 34 15 H1N1pmd09 3 4 11 16 34 15 YongHong Zhang[22] 2012 全年龄组 H1N1pmd09 9 39 35 49 98 50 Yongkun Chen[23] 2017 全年龄组 Influenza B 50 130 84 27 57 19 Zhongfang Wang[24] 2014 成人 H7N9 3 7 6 24 49 20 如表 2所示,整体而言7篇研究均获得6颗*以上(满分9颗*),研究质量可以接受。5篇研究的正常对照采用千人基因组中国人群的数据库,无法判断其基因检测的方法与病例一致,因此“病例和对照组是否采用的相同测量方法”一项均未赋*。也正是由于此类对照无法进行混杂因素的调整,可比性均为1颗*(满分2颗*)。第一篇及第三篇研究的病例量少,且未明确描述其抽样的方式,未评估其对总体的代表性,“病例的代表性”一项均未赋*。
表 2 研究纳入文献的质量评价Table 2. Quality evaluation of the included studies2.2 合并效应值定量分析结果
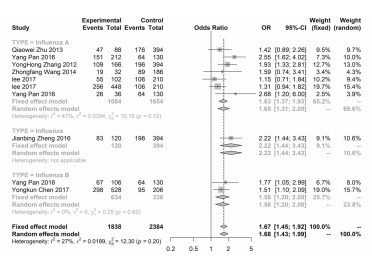
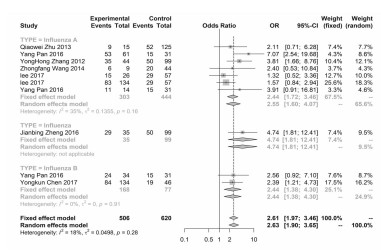
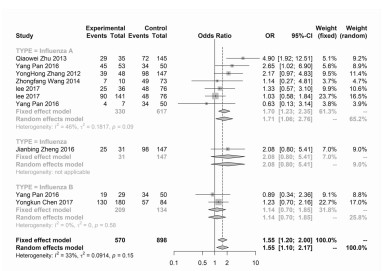
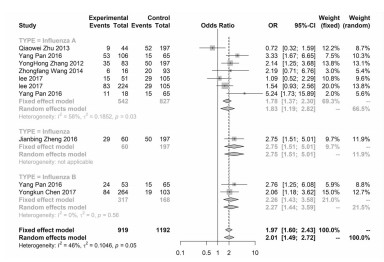
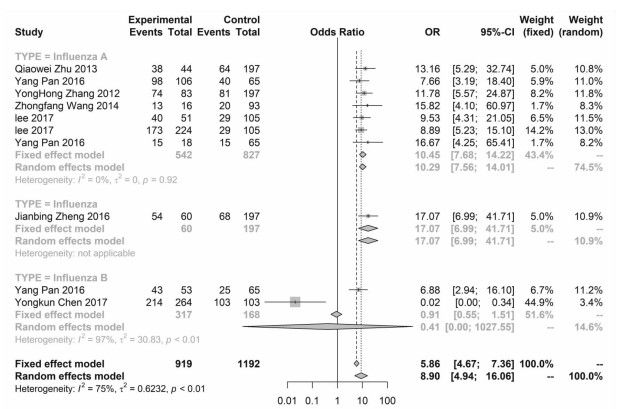
本研究将流感病例与正常人群进行比较分析,以探索不同基因型别对流感病毒感染中国人群易感性。本研究发现IFITM3基因的rs12252多态性与中国人群流感病毒易感性存在关联,rs12252 T>C多态性是流感病毒感染易感性的危险因素,见表 3。等位基因模型结果显示,携带rs12252 C等位基因位点的中国人群流感易感性是携带rs12252 T等位基因的1.67倍,95%CI为1.45~1.92,见图 2。携带rs12252 CC基因型的中国人群的流感易感性是携带rs12252 TT基因型的2.61倍,95%CI为1.97-3.46,见图 3,携带rs12252 TC基因型的中国人群的流感易感性是携带rs12252 TT基因型的1.55倍,95% CI为1.20~2.00,见图 4。但是该模型结果在乙型流感亚组分析无统计学差异(OR=1.14, 95% CI=0.7~1.85)。携带rs12252 CC基因型的中国人群的流感易感性是非携带CC基因型的2.01倍,95% CI为1.49~2.72,如图 5,非rs12252 TT基因型的中国人群的流感易感性是携带TT基因型的8.9倍,95% CI为4.94~16.06,见图 6。但是该模型结果在乙型流感亚组分析无统计学差异(OR=0.41, 95% CI=0.00~1027.55)。不论是在甲型流感病毒感染还是乙型流感病毒均存在相似的结果,即中国人群中rs12252 C等位基因更容易发生流感,见表 3。
表 3 rs12252基因多态性与中国人群流感易感性的关系(流感组vs正常人群组)Table 3. Relationship between rs12252 polymorphism and influenza susceptibility in Chinese population (influenza group vs normal group)病毒亚型 遗传模型 研究数目 OR值(95% CI) P值 Q值 Ph值 I2值 PEGGERa值 统计模型 流感 等位基因模型(C/T) 10 1.67(1.45~1.92) < 0.001 12.300 0.197 26.83 0.427 固定 流感 共显性模型1 (CC/TT) 10 2.61(1.97~3.46) < 0.001 11.006 0.275 18.23 0.549 固定 流感 共显性模型2 (TC/TT) 10 1.55(1.20~2.00) 0.001 13.339 0.148 32.53 0.837 固定 流感 隐性模型(CC/TC+TT) 10 2.01(1.49~2.72) < 0.001 16.751 0.053 46.27 0.567 随机 流感 显性模型(CC+TC/TT) 10 8.9(4.94~16.06) < 0.001 36.213 <0.001 75.15 0.889 随机 甲型流感 等位基因模型(C/T) 7 1.63(1.37~1.93) < 0.001 10.146 0.119 40.86 0.547 固定 甲型流感 共显性模型1 (CC/TT) 7 2.44(1.72~3.46) < 0.001 9.291 0.158 35.42 0.549 固定 甲型流感 共显性模型2 (TC/TT) 7 1.71(1.06~2.76) 0.001 11.053 0.087 45.71 0.771 随机 甲型流感 隐性模型(CC/TC+TT) 7 1.83(1.38~2.30) 0.006 14.358 0.026 58.21 0.642 随机 甲型流感 显性模型(CC+TC/TT) 7 10.45(7.68~14.22) < 0.001 2.047 0.915 0.00 0.134 固定 乙型流感 等位基因模型(C/T) 2 1.58(1.20~2.08) 0.001 0.251 0.617 0.00 - 固定 乙型流感 共显性模型1 (CC/TT) 2 2.44(1.38~4.30) 0.002 0.012 0.911 0.00 - 固定 乙型流感 共显性模型2 (TC/TT) 2 1.14(0.70~1.85) 0.610 0.314 0.575 0.00 - 固定 乙型流感 隐性模型(CC/TC+TT) 2 2.26(1.43~3.58) 0.001 0.345 0.557 0.00 - 固定 乙型流感 显性模型(CC+TC/TT) 2 0.41(0.00~1027.55) 0.823 28.748 < 0.001 96.52 随机 注:a文献数少于3篇将不检验发表偏倚。 2.3 异质性分析结果
从表 2中可以看到,本次研究仅仅全流感病毒及乙型流感病毒亚组中的显性模型中I2超过了60%, 说明纳入的7篇文章同质性相对较好。虽然有个别基因模型组提示存在异质性(Ph < 0.1), 但是I2均未超过70%,采用随机效应模型仍然可以得出相对可信的合并效应值。
2.4 敏感性分析结果
本次研究通过逐一删除单项文献,以判断所删除的文献对合并效应值的影响。结果发现,删除任何一个单篇研究均未对总的合并效应值产生明显影响,说明我们的研究结果是稳定的。本研究的森林图同时给出了固定效应模型和随机效应模型下的结果,绝大多数的亚组分析结果两模型相近似,且未出现固定效应模型和随机效应模型统计学差异相反的结果,间接说明了本研究的结果比较稳健。
同时对该研究方向所需的样本含量开展了基于研究项目的试验序贯分析。相关参数设置为:Ⅰ类错误α=0.05,Ⅱ类错误β=0.1,把握度为90%,流感组C等位基因的平均携带率为60%,对照组C等位基因的平均携带频率为50%。序贯分析的计算结果中,IFITM3基因rs12252位点多态性与中国人群流感易感性的研究方向若要达到稳定阳性结果需要的样本量为697例,见图 7。本研究方向共有7篇文章,共计样本含量1 682例,满足了预计样本含量的需求。同时,本研究发现累积Z-score已跨越序贯分析的监测线(Monitor Boundary)。以上说明到目前为止发表的IFITM3基因rs12252位点多态性与中国人群流感易感性的研究样本量已经足够满足该方向的样本量要求。
2.5 发表偏倚分析结果
如果所纳入研究数大于2篇,本研究采用Egger线性回归模型来定性地评估潜在的发表偏倚。发表偏倚分析结果见表 2,在任何亚组中均未发现发表偏倚(PEGGER均大于0.1)。
3. 讨论
流感病毒是引起人类呼吸道感染的病毒之一,对公众健康构成严重威胁。流感病毒含有两个重要的表面抗原,分别是血凝素(hemagglutinin protein, HA)和神经氨酸酶(neuraminidase, NA),血凝素抗原性的不断改变是导致流感反复流行的基本因素。接种疫苗对未受感染的人群有良好的预后效果,但会随着病毒抗原性变异幅度的增大而降低,而抗病毒药物则是全球防治流行的一个重要因素。近年来有报告[25]显示,现有针对流感病毒的抗病毒药物由于出现耐药毒株而变得无效。对抗流感的另一种重要策略是了解病毒-人体细胞间的相互作用,从病毒进入人体和复制时所必需的细胞因子着手,这是一个新的对抗流感的途径[26]。
有研究[27]称,胆固醇-25-羟化酶可在病毒感染的早期发挥抗病毒作用,其主要是借助羟基胆甾醇阻断病毒和宿主细胞间的膜融和过程,而IFITM蛋白家族是唯一一个在病毒感染后直接抑制病毒与宿主细胞膜融和的蛋白。人类的IFITM蛋白家族由四个成员组成:IFITM1、IFITM2、IFITM3和IFITM5,这些蛋白已被证明是流感病毒感染的有效抑制剂。IFITM蛋白家族可抑制多种病毒,如人类免疫缺陷病毒(human immunodeficiency virus,HIV)、登革热病毒、呼肠孤病毒、重症急性呼吸综合征冠状病毒(severe acute respiratory syndrome,SARS)、丝状病毒等[28-34]。因IFITM3主要在晚期内体和溶酶体中聚集,受IFITM表达影响最大的病毒是那些需要转运到亚细胞器来侵入细胞的病毒[30, 35]。虽然这似乎是抑制病毒的一个共同特征,但IFITM家族成员在他们所抑制的病毒中表现出选择性。一方面,IFITM3对甲型流感病毒有较高的抵抗力。相反,丙型肝炎病毒感染受IFITM1抑制,而IFITM3[32]则无抑制作用。在转基因小鼠模型中,一种降低IFITM3基因表达的人长非编码RNA增强了流感病毒的复制[36]。对缺乏IFITM3的小鼠进行的体内研究表明,与野生型动物相比,基因敲除小鼠的流感感染严重程度明显增加,在低致病性流感病毒的攻击下,敲除小鼠表现出暴发性病毒性肺炎,再现了1918西班牙高致病性流感病毒株[37]所造成的疾病状况。
一项在中国人群中进行的随访研究[22]发现,IFITM3基因CC基因型比CT和TT基因型更容易导致严重感染。有研究[38]证实,IFITM3基因rs12252位点多态性与人群对流感易感性有关,不过仍有研究[16]显示IFITM3基因rs12252位点多态性与人群对流感易感性无关,仅与患流感的严重程度有关。这一研究结果的不一致,导致了人们对IFITM3基因与人群对流感易感性的关系产生了怀疑。导致结果不一致的原因分析如下:一是样本量的大小,这可能导致假阳性或假阴性结果的主要原因;二是抽样误差,如种族差异;三是不同的流感病毒株在疾病程度和感染程度上的差异。本研究通过采用Meta分析的方法,综合定量分析IFITM3基因与人群流感易感性的关系,其优点是可通过增加样本含量,来降低因随机误差所造成的差异,增加结论的可信度。
本文研究结果发现,IFITM3基因的rs12252多态性与中国人群流感病毒易感性存在关联,rs12252 T>C多态性是流感病毒感染易感性的危险因素;不论是甲型流感病毒感染还是乙型流感病毒感染均存在相似的结果,即中国人群中rs12252 C等位基因更容易发生流感。以往研究[31]显示,IFITM3的表达与甲型和乙型流感感染都具有明显的关联,但这一发现均没有较大的临床研究来支持这一说法。本研究通过系统评价所有研究结果,进一步证明了IFITM3基因与中国人群流感易感性之间的关系。
综上所述,IFITM3基因rs12252多态性是流感病毒感染易感性的危险因素,中国人群中rs12252 C等位基因更容易发生流感。由于C等位基因在中国人群中出现的频率更高,中国人群在流感季节性流行或大流行时感染流感的机会将大大增加。鉴于此,该部分人群应尽早接种疫苗、早预防、早治疗,可有效降低疾病负担,减轻病痛。此外,IFITM3因其具有抗病毒的作用,可依此为药物靶点,研发新药物,为流感病毒感染者提供更为有效的药物治疗方法。
-
表 1 研究纳入文献的基本信息
Table 1. Basic information of the included studies
作者 发表年 研究对象 流感型别 流感病例基因分型 正常人群基因分型 TT TC CC TT TC CC Jianbing Zheng[19] 2016 儿童 未分型 6 25 29 49 98 50 Lee[10] 2017 成人 H7N9 11 25 15 28 48 29 H1N1pmd09 51 90 83 28 48 29 Qiaowei Zhu[20] 2013 儿童 Influenza 6 29 9 73 72 52 Yang Pan[21] 2016 全年龄组 H3N2 8 45 53 16 34 15 Yamagata 10 19 24 16 34 15 H1N1pmd09 3 4 11 16 34 15 YongHong Zhang[22] 2012 全年龄组 H1N1pmd09 9 39 35 49 98 50 Yongkun Chen[23] 2017 全年龄组 Influenza B 50 130 84 27 57 19 Zhongfang Wang[24] 2014 成人 H7N9 3 7 6 24 49 20 表 2 研究纳入文献的质量评价
Table 2. Quality evaluation of the included studies
表 3 rs12252基因多态性与中国人群流感易感性的关系(流感组vs正常人群组)
Table 3. Relationship between rs12252 polymorphism and influenza susceptibility in Chinese population (influenza group vs normal group)
病毒亚型 遗传模型 研究数目 OR值(95% CI) P值 Q值 Ph值 I2值 PEGGERa值 统计模型 流感 等位基因模型(C/T) 10 1.67(1.45~1.92) < 0.001 12.300 0.197 26.83 0.427 固定 流感 共显性模型1 (CC/TT) 10 2.61(1.97~3.46) < 0.001 11.006 0.275 18.23 0.549 固定 流感 共显性模型2 (TC/TT) 10 1.55(1.20~2.00) 0.001 13.339 0.148 32.53 0.837 固定 流感 隐性模型(CC/TC+TT) 10 2.01(1.49~2.72) < 0.001 16.751 0.053 46.27 0.567 随机 流感 显性模型(CC+TC/TT) 10 8.9(4.94~16.06) < 0.001 36.213 <0.001 75.15 0.889 随机 甲型流感 等位基因模型(C/T) 7 1.63(1.37~1.93) < 0.001 10.146 0.119 40.86 0.547 固定 甲型流感 共显性模型1 (CC/TT) 7 2.44(1.72~3.46) < 0.001 9.291 0.158 35.42 0.549 固定 甲型流感 共显性模型2 (TC/TT) 7 1.71(1.06~2.76) 0.001 11.053 0.087 45.71 0.771 随机 甲型流感 隐性模型(CC/TC+TT) 7 1.83(1.38~2.30) 0.006 14.358 0.026 58.21 0.642 随机 甲型流感 显性模型(CC+TC/TT) 7 10.45(7.68~14.22) < 0.001 2.047 0.915 0.00 0.134 固定 乙型流感 等位基因模型(C/T) 2 1.58(1.20~2.08) 0.001 0.251 0.617 0.00 - 固定 乙型流感 共显性模型1 (CC/TT) 2 2.44(1.38~4.30) 0.002 0.012 0.911 0.00 - 固定 乙型流感 共显性模型2 (TC/TT) 2 1.14(0.70~1.85) 0.610 0.314 0.575 0.00 - 固定 乙型流感 隐性模型(CC/TC+TT) 2 2.26(1.43~3.58) 0.001 0.345 0.557 0.00 - 固定 乙型流感 显性模型(CC+TC/TT) 2 0.41(0.00~1027.55) 0.823 28.748 < 0.001 96.52 随机 注:a文献数少于3篇将不检验发表偏倚。 -
[1] Tanushree D, Amita J. Influenza virus: a brief overview[J]. Proc Natl Acad Sci Sect B Biol Sci, 2012, 82(1): 111-121. DOI: 10.1007/s40011-011-009-6. [2] Fukuyama S, Kawaoka Y. The pathogenesis of influenza virus infections: the contributions of virus and host factors[J]. Curr Opin Immunol, 2011, 23(4): 481-486. DOI: 10.1016/j.coi.2011.07.016. [3] Tscherne DM, Garcia-Sastre A. Virulence determinants of pandemic influenza viruses[J]. J Clin Invest, 2011, 121(1): 6-13. DOI: 10.1172/JCI44947. [4] Wellington D, Laurenson-Schafer H, Abdel-Haq A, et al. IFITM3: how genetics influence influenza infection demographically[J]. Biomed J, 2019, 42(1): 19-26. DOI: 10.1016/j.bj.2019.01.004. [5] Desai TM, Marin M, Chin CR, et al. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion[J]. PLoS Pathog, 2014, 10(4): e1004048. DOI: 10.1371/journal.ppat.1004048. [6] Wakim LM, Gupta N, Mintern JD, et al. Enhanced survival of lung tissue-resident memory CD8(+) T cells during infection with influenza virus due to selective expression of IFITM3[J]. Nat Immunol, 2013, 14(3): 238-245. DOI: 10.1038/ni.2525. [7] Brass AL, Huang IC, Benita Y, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, west nile virus, and dengue virus[J]. Cell, 2009, 139(7): 1243-1254. DOI: 10.1016/j.cell.2009.12.017. [8] Randolph AG, Yip WK, Allen EK, et al. Evaluation of IFITM3 rs12252 association with severe pediatric influenza infection[J]. J Infect Dis, 2017, 216(1): 14-21. DOI: 10.1093/infdis/jix242. [9] Williams DE, Wu WL, Grotefend CR, et al. IFITM3 polymorphism rs12252-C restricts influenza A viruses[J]. PLoS One, 2014, 9(10): e110096. DOI: 10.1371/journal.pone.0110096. [10] Lee N, Cao B, Ke C, et al. IFITM3, TLR3, and CD55 gene SNPs and cumulative genetic risks for severe outcomes in Chinese patients with H7N9/H1N1pdm09 Influenza[J]. J Infect Dis, 2017, 216(1): 97-104. DOI: 10.1093/infdis/jix235. [11] David S, Correia V, Antunes L, et al. Population genetics of IFITM3 in portugal and central africa reveals a potential modifier of influenza severity[J]. Immunogenetics, 2018, 70(3): 169-177. DOI: 10.1007/s00251-017-1026-2. [12] Mehrbod P, Eybpoosh S, Fotouhi F, et al. Association of IFITM3 rs12252 polymorphisms, BMI, diabetes, and hypercholesterolemia with mild flu in an Iranian population[J]. Virol J, 2017, 14(1): 218. DOI: 10.1186/s12985-017-0884-4. [13] Gaio V, Nunes B, Pechirra P, et al. Hospitalization risk due to respiratory illness associated with genetic variation at IFITM3 in patients with influenza A(H1N1)pdm09 infection: a case-control study[J]. PLoS One, 2016, 11(6): e0158181. DOI: 10.1371/journal.pone.0158181. [14] López-Rodríguez M, Herrera-Ramos E, Solé-Violán J, et al. IFITM3 and severe influenza virus infection, no evidence of genetic association[J]. Eur J Clin Microbiol Infect Dis, 2016, 35(11): 1811-1817. DOI: 10.1007/s10096-016-2732-7. [15] Mills TC, Rautanen A, Elliott KS, et al. IFITM3 and susceptibility to respiratory viral infections in the community[J]. J Infect Dis, 2014, 209(7): 1028-1031. DOI: 10.1093/infdis/jit468. [16] Kim YC, Jeong BH. No Correlation of the disease severity of influenza A virus infection with the rs12252 polymorphism of the interferon-induced transmembrane protein 3 gene[J]. Intervirology, 2017, 60(1-2): 69-74. DOI: 10.1159/000479087. [17] Carter TC, Hebbring SJ, Liu J, et al. Pilot screening study of targeted genetic polymorphisms for association with seasonal influenza hospital admission[J]. J Med Virol, 2018, 90(3): 436-446. DOI: 10.1002/jmv.24975. [18] Wetterslev J, Thorlund K, Brok J, et al. Trial sequential analysis may establish when firm evidence is reached in cumulative meta-analysis[J]. J Clin Epidemiol, 2008, 61(1): 64-75. DOI: 10.1016/j.jclinepi.2007.03.013. [19] 郑建斌, 张剑珲, 陶建平, 等. 干扰素诱导跨膜蛋白3基因多态性与小儿流感严重程度相关性研究[J]. 生物医学工程学进展, 2016, 37(2): 66-69. DOI: 10.3969/j.issn.1674-1242.2016.02.002.Zheng JB, Zhang JH, Tao JP, et al. Association of interferon inducible transmembrane protein 3 gene polymorphism with the severity of influenza in children[J]. Progress in Biomedical Engineering, 2016, 37(2): 66-69. DOI: 10.3969/j.issn.1674-1242.2016.02.002. [20] 朱乔伟. 干扰素诱导的跨膜蛋白3基因rs12252 T/C多态性与儿童流感遗传易患性和严重性的相关性研究[D]. 广州: 广州医学院, 2013.Zhu QW. Correlation of rs12252 T/C polymorphism in the interferon induced transmembrane protein 3 gene and genetic susceptibility and severity in children with influenza[D]. Guangzhou: Guangzhou Medical College, 2013. [21] 潘阳, 杨鹏, 张奕, 等. 干扰素诱导跨膜蛋白3 rs12252多态性与乙型流感病毒感染相关性研究[J]. 国际病毒学杂志, 2016, 23(6): 364-367. DOI: 10.3760/cma.j.issn.1673-4092.2016.06.002.Pan Y, Yang P, Zhang Y, et al. The association between IFITM3 rs12252 polymorphism and influenza B virus infection[J]. Int J Virol, 2016, 23(6): 364-367. DOI: 10.3760/cma.j.issn.1673-4092.2016.06.002. [22] Zhang YH, Zhao Y, Li N, et al. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals[J]. Nat Commun, 2013, 4: 1418. DOI: 10.1038/ncomms2433. [23] 陈永坤, 朱闻斐, 郭俊峰, 等. 干扰素诱导跨膜蛋白3 rs12252单核苷酸多态性与乙型流感临床严重程度的相关性研究[J]. 病毒学报, 2017, 33(3): 389-393. DOI: 10.13242/j.cnki.bingduxuebao.003161.Chen YK, Zhu WF, Guo JF, et al. The association between IFITM3 rs12252 gene polymorphism and severity of influenza B virus infection[J]. Chin J Virol, 2017, 33(3): 389-393. DOI: 10.13242/j.cnki.bingduxuebao.003161. [24] Wang ZF, Zhang AL, Wan YM, et al. Early hypercytokinemia is associated with interferoninduced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection[J]. PNAS, 2014, 111(2): 769-774. DOI: 10.1073/pnas.1321748111. [25] 侯志飞, 赵学森. 干扰素诱导跨膜蛋白抗病毒机制研究进展[J/CD]. 中华实验和临床感染病杂志(电子版), 2018, 12(3): 212-215. DOI: 10.3877/cma.j.issn.1674-1358.2018.03.002.Hou ZF, Zhao XS. Advances in antiviral study on interferon-induced transmembrane protein[J/CD]. Chin J Exp Clin Infect Dis (Electronic Edition), 2018, 12(3): 212-215. DOI: 10.3877/cma.j.issn.1674-1358.2018.03.002. [26] Prabhu SS, Chakraborty TT, Kumar N, et al. Association between IFITM3 rs12252 polymorphism and influenza susceptibility and severity: a meta-analysis[J]. Gene, 2018(674): 70-79. DOI: 10.1016/j.gene.2018.06.070. [27] Brass AL, Huang IC, Benita Y, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, west nile virus, and dengue virus[J]. Cell, 2009, 139(7): 1243-1254. DOI: 10.1016/j.cell.2009.12.017. [28] Schoggins JW, Wilson SJ, Panis M, et al. A diverse range of gene products are effectors of the type I interferon antiviral response[J]. Nature, 2011, 472(7344): 481-485. DOI: 10.1038/nature09907. [29] Chen L, Borozan I, Feld J, et al. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection[J]. Gastroenterology, 2005.128(5): 1437-1444. DOI: 10.1053/j.gastro.2005.01.059. [30] Huang IC, Bailey CC, Weyer JL, et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus[J]. PLoS Pathog, 2011, 7(1): e1001258. DOI: 10.1371/journal.ppat.1001258. [31] Wilkins C, Woodward J, Lau DT, et al. IFITM1 is a tight junction protein that inhibits hepatitis C virus entry[J]. Hepatology, 2013, 57(2): 461-469. DOI: 10.1002/hep.26066. [32] Anafu AA, Bowen CH, Chin CR, et al. Interferon-inducible transmembrane protein 3(IFITM3) restricts reovirus cell entry[J]. J Biol Chem, 2013, 288(24): 17261-17271. DOI: 10.1074/jbc.M112.438515. [33] Compton AA, Bruel T, Porrot F, et al. IFITM proteins incorporated into HIV-1 virions impair viral fusion and spread[J]. Cell Host Microbe, 2014, 16(6): 736-47. DOI: 10.1016/j.chom.2014.11.001. [34] Chan YK, Huang IC, Farzan M. IFITM proteins restrict antibody-dependent enhancement of dengue virus infection[J]. PLoS One, 2012, 7(3): e34508. DOI: 10.1371/journal.pone.0034508. [35] Feeley EM, Sims JS, John SP, et al. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry[J]. PLoS Pathog, 2011, 7(10): e1002337. DOI: 10.1371/journal.ppat.1002337. [36] Ouyang J, Zhu X, Chen Y, et al. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription[J]. Cell Host Microbe, 2014, 16(5): 616-626. DOI: 10.1016/j.chom.2014.10.001. [37] Everitt AR, Clare S, Pertel T, et al. IFITM3 restricts the morbidity and mortality associated with influenza[J]. Nature, 2012, 484(7395): 519-523. DOI: 10.1038/nature10921. [38] Pan Y, Yang P, Dong T, et al. IFITM3 Rs12252-C Variant Increases Potential Risk for Severe Influenza Virus Infection in Chianese Population[J]. Front Cell Infect Microbiol, 2017, 7: 294. DOI: 10.3389/fcimb.2017.00294. 期刊类型引用(2)
1. 李世泽,梁红卫,赵迪宏. IFITM3、BTBD7、PEA3在非小细胞肺癌中的表达及临床意义. 吉林医学. 2024(01): 16-21 .  百度学术
百度学术2. 王继鹏,李朋玲,陈胜. Pokemon、miR-451、IFITM1在非小细胞肺癌组织中的表达及与病理特征的相关性. 中国老年学杂志. 2023(09): 2079-2083 .  百度学术
百度学术其他类型引用(1)
-






 下载:
下载:






 下载:
下载: