Multi-omics data integration molecular subtyping of lower-grade gliomas based on MOVICS clustering ensemble
-
摘要:
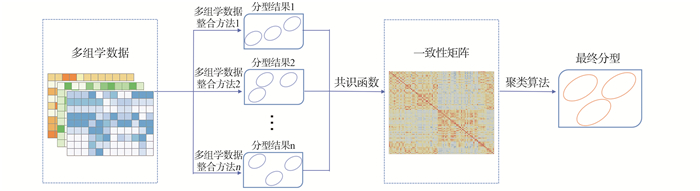
目的 探讨癌症分型中的多组学整合与可视化(multi-omics integration and visualization in cancer subtyping, MOVICS)集成聚类方法在低级别胶质瘤(lower-grade gliomas, LGG)多组学数据整合分型中的应用,识别LGG高危患者,筛选出潜在的生物标志物和重要通路。 方法 采用MOVICS方法集成LGG多组学数据的10种整合方法的分型结果,得到LGG的稳健分子分型,进一步采用Cox回归研究不同分型患者的死亡风险;针对不同分型,筛选差异表达的mRNA(DEmRNAs),miRNA(DEmiRNAs)以及差异甲基化基因(differential methylation genes, DMGs),对三者进行联合分析得到重合基因,利用GO和KEGG分析得到重合基因富集通路,进一步分析核心基因的表达水平对生存率的影响,最后对不同分型患者进行通路活性分析。 结果 LGG患者分为三型,其中,分型3患者的死亡风险是分型1的2.794倍;筛选出1 569个DEmRNAs,140个DEmiRNAs以及337个DMGs,119个重合基因富集到有统计学差异的26条GO生物功能项和7条KEGG通路;生存分析表明DNAJB14和MTUS1可能与患者生存结局相关。通路活性分析结果显示Androgen、EGFR、Trail和VEGF通路的活性在不同分型间差异有统计学意义。 结论 MOVICS聚类集成方法能够有效地对LGG患者进行分型,识别预后高风险患者,筛选出潜在生物标志物以及重要通路,为LGG患者个体化治疗策略的制定提供理论依据。 Abstract:Objective To investigate the application of multi-omics integration and visualization in cancer subtyping (MOVICS) clustering ensemble method in multi-omics data integration for lower-grade gliomas (LGG) subtyping, and high risk LGG group identification, and further screen potential biomarkers and important pathways. Methods The MOVICS method was used to integrate the subtyping results of 10 integration methods based on the LGG multi-omics data, to obtain a robust molecular subtyping of LGG patients. Cox regression analysis was carried out to evaluate the mortality risk of different patients. Differentially expressed mRNA (DEmRNAs), miRNA (DEmiRNAs) and differential methylation genes (DMGs) analyses were conducted between different subtypes. Overlapping genes among the three omics data types were used for GO term and KEGG pathway enrichment analysis. Additional analysis was conducted to identify hub genes and further evaluate their influence on patients survival outcome. Finally, pathway activity analysis between different subtypes was performed. Results LGG patients were divided into three subtypes. Patients in subtype 3 were 2.794 times more likely to die than patients in subtype 1. A total of 1 569 DEmRNAs, 140 DEmiRNAs and 337 DMGs were screened, the combined analysis genes yielded 119 genes which are regulated by mRNA, miRNA and DNA methylation and enriched 26 GO items and 7 KEGG pathways with statistical differences. Survival analysis showed that DNAJB14 and MTUS1 were significantly associated with survival outcome. Pathway activity analysis indicated that activities of Androgen, EGFR, Trail and VEGF showed significant difference between subtypes. Conclusions MOVICS classified LGG patients into three subtypes with distinct survival outcomes. Potential biomarkers, hub genes and important pathways were identified, which provided novel insights into the underlying differences between subtypes in molecular levels. These molecular signatures could offer new opportunities for individualized treatment and prevention of LGG patients. -
Key words:
- Clustering ensemble /
- Multi-omics data /
- Molecular subtype /
- Lower-grade gliomas
-
表 1 低级别胶质瘤患者分型的基本资料[ n(%)]
Table 1. Basic information on different subtypes of patients with LGG [n(%)]
项目 分型1(n=17) 分型2(n=42) 分型3(n=27) 生存时间(x±s,月) 95.953±57.614 89.157± 54.660 61.244± 55.719 年龄(岁) <40 12(70.588) 25(59.048) 15(55.556) ≥40 5(29.412) 17(40.405) 12(44.444) 性别 女性 8(47.059) 20(47.619) 12(44.444) 男性 9(52.941) 22(52.381) 15(55.556) 肿瘤分级 Ⅱ级 9(52.941) 31(73.810) 12(44.444) Ⅲ级 8(47.059) 11(26.190) 15(55.556) 生存状态 存活 11(64.706) 22(52.381) 11(40.741) 死亡 6(35.294) 20(47.619) 16(59.259) 表 2 86例低级别胶质瘤患者多变量Cox回归分析结果
Table 2. Results of multivariate Cox regression analysis of 86 patients with LGG
变量 分类 b(S.E)值 Z值 P值 HR(95% CI)值 分型 分型1 0.000 1.000 分型2 0.744(0.495) 1.503 0.133 2.104(0.797~5.551) 分型3 a 1.028(0.489) 2.102 0.036 2.794(1.072~7.285) 年龄(岁) 定量资料 0.167(0.329) 0.509 0.611 1.182(0.621~2.250) 性别 男 0.000 1.000 女 0.270(0.324) 0.835 0.404 1.311(0.695~2.473) 肿瘤分级 a 等级资料 Ⅱ级 0.000 1.000 Ⅲ级 1.557(0.346) 4.500 <0.001 4.746(2.409~9.351) 注:a P<0.05,差异有统计学意义。 -
[1] Shen F, Wu CX, Yao Y, et al. Transition over 35 years in the incidence rates of primary central nervous system tumors in Shanghai, China and histological subtyping based on a single center experience spanning 60 years[J]. Asian Pac J Cancer Prev, 2013, 14(12): 7385-7393. DOI: 10.7314/apjcp.2013.14.12.7385. [2] Xue S, Hu M, Iyer V, et al. Blocking the PD-1/PD-L1 pathway in glioma: A potential new treatment strategy[J]. J Hematol Oncol, 2017, 10(1): 81. DOI: 10.1186/s13045-017-0455-6. [3] Zhang HB, Li XS, Li YT, et al. An Immune-related signature for predicting the prognosis of lower-grade gliomas[J]. Front Immunol, 2020, 11: 603341. DOI: 10.3389/fimmu.2020.603341. [4] Xia MY, Chen HY, Chen T, et al. Transcriptional networks identify BRPF1 as a potential drug target based on inflammatory signature in primary lower-grade gliomas[J]. Front Oncol, 2021, 11: 766656. DOI: 10.3389/fonc.2021.766656. [5] Giordano TJ. The cancer genome atlas research network: a sight to behold[J]. Endocr Pathol, 2014, 25(4): 362-365. DOI: 10.1007/s12022-014-9345-4. [6] 沈思鹏, 张汝阳, 魏永越, 等. 多组学数据整合分析的统计方法研究进展[J]. 中华疾病控制杂志, 2018, 22(8): 763-765, 771. DOI: 10.16462/j.cnki.zhjbkz.2018.08.001.Shen SP, Zhang RY, Wei YY, et al. Research progress on multi-omics integrative analysis methods[J]. Chin J Dis Control Prev, 2018, 22(8): 763-765, 771. DOI: 10.16462/j.cnki.zhjbkz.2018.08.001. [7] Zhao Z, Zhang KN, Wang Q, et al. Chinese Glioma Genome Atlas (CGGA): a comprehensive resource with functional genomic data from Chinese Glioma Patients[J]. Genom Proteom Bioinf, 2021, 19(1): 1-12. DOI: 10.1016/j.gpb.2020.10.005. [8] Rappoport N, Shamir R. Multi-omic and multi-view clustering algorithms: Review and cancer benchmark[J]. Nucleic Acids Res, 2018, 46(20): 10546-10562. DOI: 10.1093/nar/gky899. [9] Alqurashi T, Wang WJ. Clustering ensemble method[J]. Springer Verlag, 2019, 10(6): 1227-1246. DOI: 10.1007/s13042-017-0756-7. [10] Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer[J]. Nat Med, 2015, 21(11): 1350-1356. DOI: 10.1038/nm.3967. [11] Strehl A, Ghosh J. Cluster ensembles---a knowledge reuse framework for combining multiple partitions[J]. J Mach Learn Res, 2002, 3: 583-617. DOI: 10.1162/153244303321897735. [12] He S, Song XY, Yang XX, et al. COMSUC: A web server for the identification of consensus molecular subtypes of cancer based on multiple methods and multi-omics data[J]. PLoS Comput Biol, 2021, 17(3): e1008769. DOI: 10.1038/s41587-019-0055-9. [13] Lu XF, Meng JL, Zhou YJ, et al. MOVICS: An R package for multi-omics integration and visualization in cancer subtyping[J]. Bioinformatics, 2020, 36(22-23): 5539-5541. DOI: 10.1093/bioinformatics/btaa1018. [14] Ramazzotti D, Lal A, Wang B, et al. Multi-omic tumor data reveal diversity of molecular mechanisms that correlate with survival[J]. Nat Commun, 2018, 9(1): 4453. DOI: 10.1038/s41467-018-06921-8. [15] Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments[J]. Stat Appl Genet Mol Biol, 2004, 3(1): 177-187. DOI: 10.2202/1544-6115.1027. [16] Ma JB, Li R, Wang J. Characterization of a prognostic four-gene methylation signature associated with radiotherapy for head and neck squamous cell carcinoma[J]. Mol Med Rep, 2019, 20(1): 622-632. DOI: 10.3892/mmr.2019.10294. [17] Dweep H, Gretz N, Sticht C. MiRWalk database for miRNA-target interactions[J]. Methods Mol Biol, 2014, 1182: 289-305. DOI: 10.1007/978-1-4939-1062-5_25. [18] Xie C, Mao XZ, Huang JJ, et al. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases[J]. Nucleic Acids Res, 2011, 39(SUPPL. 2): W316-W322. DOI: 10.1093/nar/gkr483. [19] Ashburner M, Ball CA, Blake JA, et al. Gene ontology: Tool for the unification of biology[J]. Nat Genet, 2000, 25(1): 25-29. DOI: 10.1038/75556. [20] Kanehisa M, Goto S, Sato Y, et al. KEGG for integration and interpretation of large-scale molecular data sets[J]. Nucleic Acids Res, 2012, 40(D1): D109-D114. DOI: 10.1093/nar/gkr988. [21] Schubert M, Klinger B, Klünemann M, et al. Perturbation-response genes reveal signaling footprints in cancer gene expression[J]. Nat Commun, 2018, 9(1): 1-11. DOI: 10.1038/s41467-017-02391-6. [22] Markouli M, Strepkos D, Papavassiliou AG, et al. Targeting of endoplasmic reticulum (ER) stress in gliomas[J]. Pharmacol Res, 2020, 157: 104823. DOI: 10.1016/j.phrs.2020.104823. [23] Sopha P, Kadokura H, Yamamoto Y, et al. A novel mammalian ER-located J-protein, DNAJB14, can accelerate ERAD of misfolded membrane proteins[J]. Cell Struct Funct, 2012, 37(2): 177-187. DOI: 10.1247/csf.12017. [24] Bozgeyik I, Yumrutas O, Bozgeyik E. MTUS1, a gene encoding angiotensin-Ⅱ type 2 (AT2) receptor-interacting proteins, in health and disease, with special emphasis on its role in carcinogenesis[J]. Gene, 2017, 626: 54-63. DOI: 10.1016/j.gene.2017.05.019. [25] Ranjan N, Pandey V, Panigrahi MK, et al. The tumor suppressor mtus1/atip1 modulates tumor promotion in glioma: Association with epigenetics and dna repair[J]. Cancers, 2021, 13(6): 1-21. DOI: 10.3390/cancers13061245. [26] Fu R, Ding Y, Luo J, et al. Ten-eleven translocation 1 regulates methylation of autophagy-related genes in human glioma[J]. Neuroreport, 2018, 29(9): 731-738. DOI: 10.1097/WNR.0000000000001024. [27] Hu C, Fang D, Xu HJ, et al. The androgen receptor expression and association with patient's survival in different cancers[J]. Genomics, 2020, 112(2): 1926-1940. DOI: 10.1016/j.ygeno.2019.11.005. [28] 张智峰. TRAIL及其在脑胶质瘤中的研究应用进展[J]. 国外医学: 神经病学神经外科学分册, 2002, 29(4): 363-366. DOI: 10.16636/j.cnki.jinn.2002.04.027.Zhang ZF. Progress in TRAIL and its application in glioma[J]. Foreign Med Sci: Psychiatry, 2002, 29(4): 363-366. DOI: 10.16636/j.cnki.jinn.2002.04.027. [29] Pollack IF, Erff M, Ashkenazi A. Direct stimulation of apoptotic signaling by soluble Apo2l/tumor necrosis factor-related apoptosis-inducing ligand leads to selective killing of glioma cells[J]. Clin Cancer Res, 2001, 7(5): 1362-1369. [30] 邓钢, 陈谦学. 恶性胶质瘤中的EGFR-STAT3信号通路[J]. 中国神经肿瘤杂志, 2012, 10(3): 205-208.Deng G, Chen QX. EGFR-STAT3 signal pathway in malignant glioma[J]. Chin J Neuro-Oncol, 2012, 10(3): 205-208. [31] Wang H, Wang X, Xu L, et al. Analysis of the EGFR Amplification and CDKN2A Deletion Regulated Transcriptomic Signatures Reveals the Prognostic Significance of SPATS2L in Patients with Glioma[J]. Front Oncol, 2021, 11: 713. DOI: 10.3389/fonc.2021.551160. [32] 张学新, 苏君, 常亮, 等. VEGF在低级别胶质瘤复发, 恶变过程中的表达及意义[J]. 中国误诊学杂志, 2012, 12(2): 329-330. https://www.cnki.com.cn/Article/CJFDTOTAL-ZWZX201202073.htmZhang XX, Su J, Chang L, et al. Expression and significance of VEGF in low grade glioma relapse and malignant transformation[J]. Chin J Misdiagn, 2012, 12(2): 329-330. https://www.cnki.com.cn/Article/CJFDTOTAL-ZWZX201202073.htm -





 下载:
下载: