Using molecular transmission networks to analyze the epidemiological characteristics of HIV-1 CRF01_AE in Kunming
-
摘要:
目的 通过构建昆明市HIV-1 CRF01_AE毒株的分子传播网络,分析其流行特征并观察动态流行趋势,从而为当地制定疫情防控干预措施提供科学依据。 方法 选择2015―2021年云南省传染病医院接受治疗的253例昆明市HIV-1 CRF01_AE新发感染患者作为研究对象。运用反转录巢式聚合酶链式反应成功扩增其基因序列,序列比对后导入HyPhy 2.2.4软件进行成对基因距离的计算。运用GraphPad-Prism 8.0软件确定最佳基因距离阈值,使用Cytoscope 3.7.2软件实现网络可视化。运用Network-Analyzer和MCODE(Molecular Complex Detection)工具进行网络特征分析。 结果 在0.018最佳基因距离阈值下,253个研究对象中有118个(46.64%)进入网络分析,共形成了38个分子簇,其组成大小从2到17个节点不等。网络集群主要以异性传播(51.78%)和同性传播(37.94%)为主,年龄段主要分布于20~40岁(77.47%)。网络所有节点的总链接数为226,单个节点最高链接数为10。MCODE确定了3种重要的分子簇,3个分子簇分别定义为B、C、D型,B型由17个节点和19链接数组成,为同性传播集群呈低增长状态。C和D型均由5个节点和10链接数组成,为异性传播集群且呈静止状态。 结论 昆明市HIV-1 CRF01_AE毒株分子传播网络中的分子簇具有一定特殊性和聚集性。同性传播和异性传播人群已经成为昆明市HIV-1 CRF01_AE毒株感染的两大风险群体,同时两个群体还有发生交叉传播的现象。传播网络中有一个同性传播组成的集群规模较大,其传播风险和活跃度较高,需加强对其监测,并制定针对性的干预措施对集群中的“核心人群”进行防控。 -
关键词:
- HIV-1 CRF01_AE /
- 分子传播网络 /
- 成对基因距离 /
- 分子簇
Abstract:Objective To construct the molecular transmission network of the HIV-1 CRF01_AE strain in Kunming, to analyze its epidemiological characteristics, to observe its dynamic epidemiological trends, and to provide a scientific basis for the development of local epidemic prevention and control interventions. Methods A total of 253 newly infected patients with HIV-1 CRF01_AE in Kunming who received treatment at Yunnan Infectious Disease Hospital from 2015 to 2021 were selected as study subjects. Their gene sequences were successfully amplified using RT-nested PCR (reverse transcription nested polymerase chain reaction), and the sequences were compared and imported into HyPhy 2.2.4 software for the calculation of paired gene distances. GraphPad-Prism 8.0 software was used to determine the optimal gene distance threshold, and Cytoscope 3.7.2 software was used for network visualization. Network characterization was performed using the Network Analyzer and MCODE (Molecular Complex Detection) tools. Results At an optimal genetic distance threshold of 0.018, 118 of 253 study subjects (46.64%) entered the network analysis, forming a total of 38 molecular clusters with composition sizes ranging from 2 to 17 nodes. The network clusters were mainly heterosexual (51.78%) and homosexual (37.94%), and the age group was mainly distributed between 20 and 40 years old (77.47%). The total number of links for all nodes in the network was 226; the maximum number of links for a single node was 10. MCODE identified three important molecular clusters, types B, C and D. Type B consisted of 17 nodes and 19 link counts and was a homosexually propagating cluster in a low-growth state. Types C and D both consisted of 5 nodes and 10 link counts, were heterosexually propagating clusters, and were stationary. Conclusions The molecular clusters in the molecular transmission network of the HIV-1 CRF01_AE strain in Kunming have certain specificity and aggregation. Homosexual and heterosexual transmission populations have become the two major risk groups for HIV-1 CRF01_AE strain infection in Kunming, while cross-transmission occurs in both groups. It is worth noting that there is a large cluster of same-sex transmission in the transmission network with high transmission risk and activity, which needs to be monitored. Targeted interventions should be developed to prevent and control the "core population" in the cluster. -
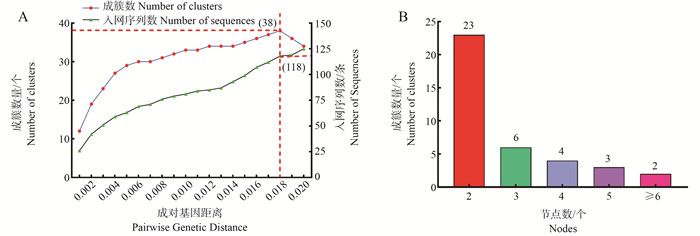
图 1 HIV-1 CRF01_AE毒株成对基因距离阈值及最佳阈值下网络成簇情况
A:为不同成对基因距离下,入网的序列数量(右纵坐标)和成簇的数量(左纵坐标)趋势图;红色虚线:成簇数量到达峰值时所对应的入网序列数和成对基因距离;B:最佳阈值为0.018时,由不同节点组成的簇的数量的频数直方图。
Figure 1. HIV-1 CRF01_AE strain pairwise gene distance thresholds and network clustering at optimal thresholds
A: trends of the number of sequences entering the network (right vertical coordinate) and the number of clusters (left vertical coordinate) with different pairwise gene distances; Red dashed line: the number of sequences entering the network and the pairwise gene distance when the number of clusters reaches the peak; B: frequency histogram of the number of clusters composed of different nodes at the optimal threshold of 0.018.
图 2 HIV-1 CRF01_AE毒株分子传播网络特征
HET:异性传播; IDUs: 静脉吸毒;A/B/C/D:分别为传播途径、年龄段传播途径和年龄段交互、传播途径和性别的分子传播网络;A1/B1/C1/D1:各组传播网络中节点的总连接数直方图;A2/B2/C2/D2:各组传播网络中节点连接数的箱式图。
Figure 2. Characterization of the molecular transmission network of HIV-1 CRF01_AE strain
HET: Heterosexual transmission; IDUs: intravenous drug use; A/B/C/D: molecular transmission network of transmission ways, age groups, age groups transmission ways plus age group interaction, and transmission ways plus gender, respectively; A1/B1/C1/D1: histograms of the total number of connections of the nodes in the transmission networks of each group; A2/B2/C2/D2: box plots of the number of connections of the nodes in the transmission networks of each group graph.
图 3 HIV-1 CRF01_AE毒株分子传播网络节点变化趋势
A: 节点的入网率趋势图;B: 同性传播集群中节点增长情况;C、D: 两个不同异性传播集群中节点增长情况。
Figure 3. Trends in node changes in the molecular transmission network of the HIV-1 CRF01_AE strain
A: trend graph of the node′s enrollment rate; B: node growth in the same-sex transmission clusters; C and D: node growth in two different opposite-sex transmission clusters.
表 1 昆明市HIV-1 CRF01_AE毒株分子传播网络基本信息
Table 1. Basic information on the molecular transmission network of HIV-1 CRF01_AE strain in Kunming
项目Projects 人数 ② Populations ② 入网人数 ② Number of clustering ② 未入网人数 ② Number of not-clustering ② χ2值value P值value 总体Total 253(100.00) 118(46.64) 135(53.36) 性别Gender 0.679 0.410 男Male 209(82.61) 95(45.45) 114(54.55) 女Female 44(17.39) 23(52.28) 21(47.72) 年龄组/岁Age group/years 0.984 ③ 0.842 < 20 6(2.37) 3(50.00) 3(50.00) 20~40 196(77.47) 90(45.92) 106(54.08) >40~60 43(17.00) 20(46.52) 23(53.48) >60 8(3.16) 5(62.50) 3(37.50) 传播途径Transmission categories 2.130 0.546 静脉吸毒Intravenous drug use 14(5.53) 4(28.57) 10(71.43) 异性传播Heterosexual transmission 131(51.78) 63(48.09) 68(51.91) 同性传播Homosexual transmission 96(37.94) 46(47.92) 50(52.08) 其他 ① Others ① 12(4.75) 5(41.67) 7(58.33) 婚姻状态Marital status 1.655 0.437 未婚Single 116(45.85) 51(43.97) 65(56.03) 已婚Married 113(44.66) 53(46.90) 60(53.10) 离异/丧偶Divorced/Widower 24(9.49) 14(58.33) 10(41.67) 采样时间/年Sampling/years 30.310 < 0.001 2015 33(13.04) 18(54.55) 15(45.45) 2016 37(14.62) 21(56.76) 16(43.24) 2017 59(23.32) 33(55.93) 26(44.07) 2018 43(17.00) 24(55.81) 19(44.19) 2019 25(9.88) 14(56.00) 11(44.00) 采样时间/年Sampling/years 2020 23(9.10) 3(13.04) 20(86.96) 2021 33(13.04) 5(15.15) 28(84.85) CD4细胞计数/(个·L-1) CD4 cell count/(cells·L-1) 6.474 0.039 < 200 71(28.06) 28(39.44) 43(60.56) 200~500 99(39.13) 56(56.57) 43(43.43) >500 83(32.81) 34(40.96) 49(59.04) 病毒载量/(拷贝·mL-1) Viral load/(copies·mL-1) 4.532 0.104 < 104 84(33.20) 39(46.43) 45(53.57) 104~105 110(43.39) 58(52.73) 52(47.27) >105 59(23.41) 21(35.59) 38(64.41) 注:CD4,CD4+T淋巴细胞。
①母婴传播或输血感染或报告不明; ②以人数(占比/%)表示; ③ Fisher确切概率法。
Notes: CD4,CD4+ T lymphocytes.
① Mother-to-child transmission or transfusion infection or report unknown; ② Number of people (proportion/%); ③ Fisher′s exact test. -
[1] Shushtari ZJ, Hosseini SA, Sajjadi H, et al. Social network and HIV risk behaviors in female sex workers: a systematic review[J]. BMC Public Health, 2018, 18(1): 1020. DOI: 10.1186/s12889-018-5944-1. [2] Billock RM, Powers KA, Pasquale DK, et al. Prediction of HIV transmission cluster growth with statewide surveillance data[J]. JAIDS J Acq Imm Def, 2019, 80(2): 152-159. DOI: 10.1097/qai.0000000000001905. [3] Fan Q, Zhang JF, Luo MY, et al. Molecular genetics and epidemiological characteristics of HIV-1 epidemic strains in various sexual risk behaviour groups in developed Eastern China, 2017-2020[J]. Emerg Microbes Infect, 2022, 11(1): 2326-2339. DOI: 10.1080/22221751.2022.2119167. [4] Aldous JL, Pond SK, Poon A, et al. Characterizing HIV transmission networks across the United States[J]. Clin Infect Dis, 2012, 55(8): 1135-1143. DOI: 10.1093/cid/cis612. [5] Brenner BG, Roger M, Moisi DD, et al. Transmission networks of drug resistance acquired in primary/early stage HIV infection[J]. AIDS, 2008, 22(18): 2509-2515. DOI: 10.1097/qad.0b013e3283121c90. [6] Gibson KM, Jair K, Castel AD, et al. A cross-sectional study to characterize local HIV-1 dynamics in Washington, DC using next-generation sequencing[J]. Sci Rep, 2020, 10: 1989. DOI: 10.1038/s41598-020-58410-y. [7] Chen M, Ma YL, Chen HC, et al. Spatial clusters of HIV-1 genotypes in a recently infected population in Yunnan, China[J]. BMC Infect Dis, 2019, 19(1): 669. DOI: 10.1186/s12879-019-4276-9. [8] Chen M, Yang L, Ma YL, et al. Emerging variability in HIV-1 genetics among recently infected individuals in Yunnan, China[J]. PLoS One, 2013, 8(3): e60101. DOI: 10.1371/journal.pone.0060101. [9] Li K, Liu ML, Chen HH, et al. Using molecular transmission networks to understand the epidemic characteristics of HIV-1 CRF08_BC across China[J]. Emerg Microbes Infect, 2021, 10(1): 497-506. DOI: 10.1080/22221751.2021.1899056. [10] Deng XM, Liu JF, Li JJ, et al. Prevalence of HIV-1 drug-resistance genotypes among unique recombinant forms from Yunnan Province, China in 2016-2017[J]. AIDS Res Hum Retroviruses, 2020, 36(5): 389-398. DOI: 10.1089/aid.2019.0041. [11] Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees[J]. Mol Biol Evol, 1993, 10(3): 512-526. DOI: 10.1093/oxfordjournals.molbev.a040023. [12] Otasek D, Morris JH, Bouças J, et al. Cytoscape Automation: empowering workflow-based network analysis[J]. Genome Biol, 2019, 20(1): 185. DOI: 10.1186/s13059-019-1758-4. [13] Halary S, Leigh JW, Cheaib B, et al. Network analyses structure genetic diversity in independent genetic worlds[J]. Proc Natl Acad Sci USA, 2010, 107(1): 127-132. DOI: 10.1073/pnas.0908978107. [14] Zhang YY, Dai J, Li ZX, et al. Using molecular network analysis to explore the characteristics of HIV-1 transmission in a China-Myanmar border area[J]. PLoS One, 2022, 17(5): e0268143. DOI: 10.1371/journal.pone.0268143. [15] Banerjee S, Baah-Acheamfour M, Carlyle CN, et al. Determinants of bacterial communities in Canadian agroforestry systems[J]. Environ Microbiol, 2016, 18(6): 1805-1816. DOI: 10.1111/1462-2920.12986. [16] 赵帅, 冯毅, 辛若雷, 等. 应用分子传播网络研究北京男男性行为者HIV-1毒株的传播特征[J]. 中国艾滋病性病, 2018, 24(3): 241-245, 306. DOI: 10.13419/j.cnki.aids.2018.03.08.Zhao S, Feng Y, Xin RL, et al. Using molecular transmission network to explore the transmission characteristics of HIV-1 among men who have sex with men in Beijing[J]. Chin J AIDS STD, 2018, 24(3): 241-245, 306. DOI: 10.13419/j.cnki.aids.2018.03.08. [17] Wertheim JO, Kosakovsky Pond SL, Forgione LA, et al. Social and genetic networks of HIV-1 transmission in New York city[J]. PLoS Pathog, 2017, 13(1): e1006000. DOI: 10.1371/journal.ppat.1006000. [18] Delva W, Leventhal GE, Helleringer S. Connecting the dots: network data and models in HIV epidemiology[J]. AIDS Lond Engl, 2016, 30(13): 2009-2020. DOI: 10.1097/QAD.0000000000001184. [19] 杨垚, 梁华悦, 钟姗妹, 等. 2018年广西崇左边境ART人群HIV-1分子网络特征分析[J]. 中国艾滋病性病, 2021, 27(6): 577-581. DOI: 10.13419/j.cnki.aids.2021.06.03.Yang Y, Liang HY, Zhong SM, et al. Molecular Network Analysis of HIV/AIDS patients receiving antiviral therapy in Chongzuo, Guangxi in 2018[J]. Chin J AIDS STD, 2021, 27(6): 577-581. DOI: 10.13419/j.cnki.aids.2021.06.03. [20] 甘梦泽, 冯毅, 邢辉. 基于分子网络方法研究HIV感染者传播特征的相关进展[J]. 中华流行病学杂志, 2019, 40(11): 1487-1491. DOI: 10.3760/cma.j.issn.0254-6450.2019.11.026.Gan MZ, Feng Y, Xing H. Progress in research on the transmission characteristics of HIV-infected persons based on molecular network method[J]. Chin J Epidemiol, 2019, 40(11): 1487-1491. DOI: 10.3760/cma.j.issn.0254-6450.2019.11.026. [21] Chen M, Ma YL, Chen HC, et al. HIV-1 genetic transmission networks among men who have sex with men in Kunming, China[J]. PLoS One, 2018, 13(4): e0196548. DOI: 10.1371/journal.pone.0196548. [22] Wertheim JO, Murrell B, Mehta SR, et al. Growth of HIV-1 molecular transmission clusters in New York city[J]. J Infect Dis, 2018, 218(12): 1943-1953. DOI: 10.1093/infdis/jiy431. [23] Mehta SR, Wertheim JO, Brouwer KC, et al. HIV transmission networks in the San diego-tijuana border region[J]. EBioMedicine, 2015, 2(10): 1456-1463. DOI: 10.1016/j.ebiom.2015.07.024. [24] Han X, Zhao B, An M, et al. Molecular network-based intervention brings us closer to ending the HIV pandemic[J]. Front Med. 2020, 14(2): 136-148. DOI: 10.1007/s11684-020-0756-y. -





 下载:
下载: